 Cytotoxic
stimuli [1a.] or Programmed cell death, via nitric oxide generation,
lead to the binding of GAPDH from its usual tetrameric form to a
dimeric form, to the protein Siah1 [1.] intracellular G-3-P [2.]
substrate [3.] protects GAPDH from S-nitrosylation [4.]. The
GAPDH-Siah interaction depends on lysine 227
[5.], in human GAPDH that interacts with a large groove [6.] of the
Siah1 dimer, that connects the GAPDH dimer to PGK in the cytoplasm.
Cytotoxic
stimuli [1a.] or Programmed cell death, via nitric oxide generation,
lead to the binding of GAPDH from its usual tetrameric form to a
dimeric form, to the protein Siah1 [1.] intracellular G-3-P [2.]
substrate [3.] protects GAPDH from S-nitrosylation [4.]. The
GAPDH-Siah interaction depends on lysine 227
[5.], in human GAPDH that interacts with a large groove [6.] of the
Siah1 dimer, that connects the GAPDH dimer to PGK in the cytoplasm.
 The
S-nitrosylation
[7.,8.] abolishes catalytic activity and confers upon GAPDH the
ability to bind to Siah [9.]. (GAPDH) is physiologically nitrosylated
at its Cys 150 residue. GAPDH (SNO-GAPDH) [10.] binds to Siah1 [11.]
by forming a protein complex. In the nucleus [12.] GAPDH is
acetylated at Lys 160 [13.] and binds to the protein
acetyltransferase p300/CBP. Under these conditions siah-1 formed a
complex with GAPDH (PDB:4O63) and localized in the nucleus of Müller
cells [14.]. GAPDH mutants [15.] that cannot bind Siah1 prevents
translocation [16.] to the nucleus to elicit neurotoxicity [17.] and
cell apoptosis.
The
S-nitrosylation
[7.,8.] abolishes catalytic activity and confers upon GAPDH the
ability to bind to Siah [9.]. (GAPDH) is physiologically nitrosylated
at its Cys 150 residue. GAPDH (SNO-GAPDH) [10.] binds to Siah1 [11.]
by forming a protein complex. In the nucleus [12.] GAPDH is
acetylated at Lys 160 [13.] and binds to the protein
acetyltransferase p300/CBP. Under these conditions siah-1 formed a
complex with GAPDH (PDB:4O63) and localized in the nucleus of Müller
cells [14.]. GAPDH mutants [15.] that cannot bind Siah1 prevents
translocation [16.] to the nucleus to elicit neurotoxicity [17.] and
cell apoptosis.[1a.] 16492755, 8769851003 [1.]16391220, [2.]19542219, 22534308, 3350006004, 19937139, [3.]22847419, [4.]15951807, [5.]20601085, [6.]16510976, 20392205005, [7.,8.]22817468006, 16505364007, [9.]16633896, [10.]16574384, [11.]20972425, [12.]19607794, [13.]18552833, [14.]19940145, [15.]23027902008, [16.]24362262, [17.]16492755.

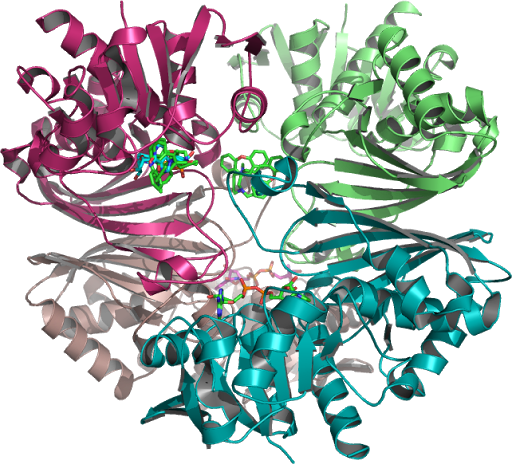 Analysis
of CGP-3466 Docking (NAD) to Human Placental GAPDH which decreases
the synthesis of pro-apoptotic proteins is N-terminally
PMID:10677844, processed to which a Rossmann NAD(P) binding fold as
seen in figure 1 is a C-terminal domain as seen on this page,
PMID:10617673, 26022259, 16510976 ...The structure is also used to
build a model of the complex between GAPDH and the E3 ubiquitin
ligase Siah1. (Purple Ribbon-1U8F_Q Figure 1.)
Analysis
of CGP-3466 Docking (NAD) to Human Placental GAPDH which decreases
the synthesis of pro-apoptotic proteins is N-terminally
PMID:10677844, processed to which a Rossmann NAD(P) binding fold as
seen in figure 1 is a C-terminal domain as seen on this page,
PMID:10617673, 26022259, 16510976 ...The structure is also used to
build a model of the complex between GAPDH and the E3 ubiquitin
ligase Siah1. (Purple Ribbon-1U8F_Q Figure 1.)

 In
the GAPDH-catalyzed reaction these intermediate metabolites
(aldolase, triose-phosphate-isomerase Glycolysis and Glyconeogenesis
(TPI)) catalyze the Glycolysis reactions, in the sequence of the ten
enzyme-catalyzed Embden-Meyerhof reactions in the metabolic
pathway. Converting phosphoglycerate mutase 1 (PGM) catalyzing the
internal steps by 2,3-BPG phosphatase to form by converting
D-glyceraldehyde 3-phosphate g3p(G3P) into 1,3-bisphosphoglycerate
(1,3-BPG) from its role as 3-Phosphoglyceric acid (3PG) in glycolysis
as the glycolytic protein GAPDH that catalyzes the first step (G3P
into 1,3-BPG) of the pathway.
In
the GAPDH-catalyzed reaction these intermediate metabolites
(aldolase, triose-phosphate-isomerase Glycolysis and Glyconeogenesis
(TPI)) catalyze the Glycolysis reactions, in the sequence of the ten
enzyme-catalyzed Embden-Meyerhof reactions in the metabolic
pathway. Converting phosphoglycerate mutase 1 (PGM) catalyzing the
internal steps by 2,3-BPG phosphatase to form by converting
D-glyceraldehyde 3-phosphate g3p(G3P) into 1,3-bisphosphoglycerate
(1,3-BPG) from its role as 3-Phosphoglyceric acid (3PG) in glycolysis
as the glycolytic protein GAPDH that catalyzes the first step (G3P
into 1,3-BPG) of the pathway.

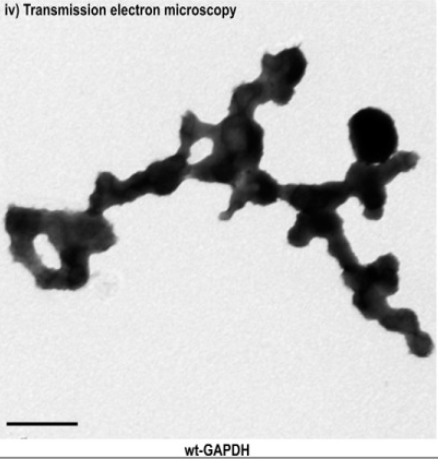 GAPDH
homotetramer was studied as represented an assembly of repeating
spherical units that harbored a distinct birefringent crystal
structure to the optic axis for the p polarization, also (r axis)
discernible via transmission electron microscopy. of the relative
amount of soluble monomeric GAPDH to G3P in the binding pocket of the
NAD(+)-binding site residue located at the active site linked to
GAPDH in Figures 5 and 6. PMID:10407144009,
25086035.
GAPDH
homotetramer was studied as represented an assembly of repeating
spherical units that harbored a distinct birefringent crystal
structure to the optic axis for the p polarization, also (r axis)
discernible via transmission electron microscopy. of the relative
amount of soluble monomeric GAPDH to G3P in the binding pocket of the
NAD(+)-binding site residue located at the active site linked to
GAPDH in Figures 5 and 6. PMID:10407144009,
25086035.
 Another
model building studie indicates that a structure obtained where
glyceraldehyde 3-phosphate PDB:3CMC_Q binds in the P(s) pocket of the
natural substrate G3P phosphorylating GAPDH (PDB:1U8F_Q) at the
catalytic cysteine residue site. To define the conditions suitable
for affinity for the cosubstrate, the isolation and accumulation of
the intermediate metabolites per G3P monomer found in Figure 8 of the
equivalent Glc-3-P structure in the binding pocket of the
NAD(+)-binding site residue located at the active site linked to
GAPDH. PMID:19542219,
22534308
Another
model building studie indicates that a structure obtained where
glyceraldehyde 3-phosphate PDB:3CMC_Q binds in the P(s) pocket of the
natural substrate G3P phosphorylating GAPDH (PDB:1U8F_Q) at the
catalytic cysteine residue site. To define the conditions suitable
for affinity for the cosubstrate, the isolation and accumulation of
the intermediate metabolites per G3P monomer found in Figure 8 of the
equivalent Glc-3-P structure in the binding pocket of the
NAD(+)-binding site residue located at the active site linked to
GAPDH. PMID:19542219,
22534308
 Correctly
known binding sites on ((GAPD/NAD)) structures, polar spheres of the
binding catalytic pocket that corresponds to G3P (glyceraldehyde
3-phosphate) aligned to the holographical structure nonbounded
spheres (salmon color), these apoenzymes together with the
cofactor(s) Cys 151, 152 which corresponds as below the Ps pocket of
G3P, on the Green ribbon required for cofactor activity. Together
with eliminated crystallographic waters and other possible spheres,
these are at least one atom of a amino acid residue in contact with
at least one alpha sphere of one binding pocket on the holo protein
NAD structure 1U8F_Q needed to align holo and apo structures included
in this data set with G3P (PDB:3CMC_Q) was tested only on holo
structure (NAD), obtained via Pea Green spheres aligned to 1U8F_Q
ribbons/ligand structure which provide structural recognition
insights into the biological 1U8F-Q assembly this includes 29
asymmetric units of its dimeric form, along the tetrameric 1U8F
biological forms axis. PMID:9461340010
Correctly
known binding sites on ((GAPD/NAD)) structures, polar spheres of the
binding catalytic pocket that corresponds to G3P (glyceraldehyde
3-phosphate) aligned to the holographical structure nonbounded
spheres (salmon color), these apoenzymes together with the
cofactor(s) Cys 151, 152 which corresponds as below the Ps pocket of
G3P, on the Green ribbon required for cofactor activity. Together
with eliminated crystallographic waters and other possible spheres,
these are at least one atom of a amino acid residue in contact with
at least one alpha sphere of one binding pocket on the holo protein
NAD structure 1U8F_Q needed to align holo and apo structures included
in this data set with G3P (PDB:3CMC_Q) was tested only on holo
structure (NAD), obtained via Pea Green spheres aligned to 1U8F_Q
ribbons/ligand structure which provide structural recognition
insights into the biological 1U8F-Q assembly this includes 29
asymmetric units of its dimeric form, along the tetrameric 1U8F
biological forms axis. PMID:9461340010
 (Figure
8.) These are the results without the liquid chromatography coupled
mass spectrometer, that are known 3D products by two-dimensional
sequence analyses with the STRAP alignment tools data sets and which
may have any effect on the functions of further analysis involved in
more ordered results than this study attempts to show, of the
analysis that may be included are identified separated into multiple
gradients here in these paired graphs. Therefore in the present work
to uncover the exact coincidence of 1U8F_R and 4I7D_C, the 3D
coordinates of GAPDH (PDB:1U8F_Q) to the protein Siah1 4I7D were not
presenting when subjected to STRAP alignment this apparent
discrepancy (Figure 1.) was partially resolved by a (Figure 7)
rendering from a more reactive native GAPDH_R homotetramer model.
(Figure
8.) These are the results without the liquid chromatography coupled
mass spectrometer, that are known 3D products by two-dimensional
sequence analyses with the STRAP alignment tools data sets and which
may have any effect on the functions of further analysis involved in
more ordered results than this study attempts to show, of the
analysis that may be included are identified separated into multiple
gradients here in these paired graphs. Therefore in the present work
to uncover the exact coincidence of 1U8F_R and 4I7D_C, the 3D
coordinates of GAPDH (PDB:1U8F_Q) to the protein Siah1 4I7D were not
presenting when subjected to STRAP alignment this apparent
discrepancy (Figure 1.) was partially resolved by a (Figure 7)
rendering from a more reactive native GAPDH_R homotetramer model.
References: