Triosephosphate
isomerase (TPI, EC 5.3.1.1)
(§,
‡) is essential to glycolysis, catalyzes the fifth step in the
glycolysis pathway the reversible conversion of dihydroxyacetone
phosphate (DHAP) into glyceraldehyde-3-phosphate. TPI is a homodimer
formed by two identical dimeric molecules of a single structural
locus : 12p13.31.
TPI has only 1 functional gene with a molecular mass of 29 kDa, that
after refinement are products of a distinct single
structural locus. The variant phenotype of identical subunits are
expressed in both red cells and circulating lymphocytes,
catalyzing the interconversion of one of the two products breakdown
by reversible
conversion. The TPI substrate by deprotonation
the transition state reaction of dihydroxyacetone phosphate (DHAP)
substrate yields one product of the glycolytic pathway, is a trend*
(Kcat) that persists creating the initial complex
microcompartmentation of TPI to give (G3P)
glyceraldehyde-3-phosphate which seems to be the isomerase*
activity, release is slower than its conversion to DHAP in normal
and TPI deficient cells. TIM
with its natural substrates has not
been
(•)
crystalized**.
TPI is a dimeric enzyme and contains 7 exons interrupted by six
introns.
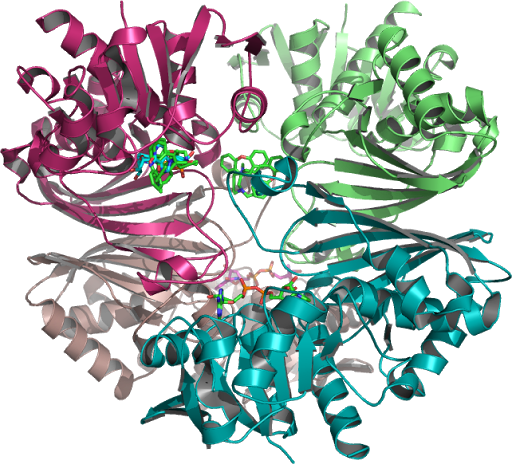
The crystallographic structure of (HsTPI) human triosephosphate isomerase PDB:1HTI is one dimer per asymmetric unit subunit 1 and subunit 2 are in the open and closed conformations in the 3-dimensional asymmetric space group P 2(1) which is specific to the Monoclinic with minimization on the entire structure in the presence of substrate analogues and its surrounding residues supporting possible regions targeted for drug design.
TPI
deficiency (TPID) a disorder of glycolysis, occurring in haplotypes
of specific alleles heterogeneous to clinical TPI-deficiency,
with a rare homozygous deficiency
the resulting genetic
defect is the cause of a null variant incompatible with life
by abnormally high levels
of DHAP
which degrades spontaneously into the toxic (MG)
methylglyoxal,
due to deamidation
of asparagine (Asn15-71)
to form
aspartic and glutamic acid. Loop
6
plays a role in preventing the breakdown yield of methylglyoxal
(fMG)
one of the of the three products of enzyme-bound enediol(ate)
phosphate,
towards elimination
of (fMG)
inorganic phosphate. TPI deficiency is due to the common aberrant
dimerization (or the dissociation into inactive monomers) of mutation
TPI 1591C,
encoding a Glu104-to-Asp
(glutamate-to-aspartate) substitution in the TPI variant
found in cases of hemolytic anemia coupled
with neurodegeneration,
the Glu104-to-Asp
substitution is the most common
disease allele inherited, when compared to wild-type TPI's three
(residues from the same subunit)
similar but not identical interactions between the inhibitor and
catalytic residues, Glu 167
(or 165)
forms a stable dimer and provides the rationale
for production of structurally normal enzyme in humans, the E104D
mutation, provides the amyloid-resistant
structure of human triosephosphate isomerase (HsTPI).
Water-protein
molecules join
two catalytically active monomers which is only in its dimeric form,
as monomers
of TIM are not functional. Within a hydrophobic catalytic pocket
of the native enzymes the binding and catalysis of TPIs in
hemolysates,
bind to the red
cell
membrane. Molecular modeling using the human crystal structure of
TPI
was performed to determine how these mutations could affect enzyme
structure and function. The Amyloid secondary
structure autoepitopes antigen-driven
mechanism works toward recovery of the anti-triosephosphate
isomerase mutant TPI peptide**
antigens. This is the scheme
that allows function-enhancing stability most significantly, the
catalysis for deprotonation of DHAP or vice-versa GAP substrates of
the TIM-barrel relative to TPI toward turnover of two-part substrate
glycolaldehyde / phosphite dianion {GA + HPO32* the transition state
for this enolising enzyme substrate pieces.} Km/obsd*
group of the whole GAP substrate and H95
(loop 4)
is also optimal for small mutational changes in or reflects its
compatibility with amino acid residues which stabilizes the
enediolate
intermediate (GA/HPO)
activity from change in the products scheme
(a proton transfer mechanism)
DHAP/G3P
or interconversion of these intermediates.

Closed (activated for catalysis) of optimal WT (TPI) molecular modeling PDB 1HTI_B using the human crystal structure of TPI human triosephosphate isomerase (HsTPI) conformation 1hti_b, calculated to the incidence residue Water-protein molecules and the protein cage that interacts within a hydrophobic catalytic pocket isolated and examined which coded for human triose-phosphate isomerase. [EC: 5.3.1.1]….
The
active flexible site loop must open
before product release,
unliganded in trypanosomal Tb-TIM
glycerol phosphate ester to liganded Glu167
in the catalytic cycle and the enzymes substrate transition state
between open
and closed
to protect the substrate for the turnover of DHAP and G3P (GAP) the
natural substrates, and inhibiting the formation of a toxic
by-product in the absence
of this equilibration reactions between dihydroxyacetone phosphate
and glyceraldehyde 3-phosphate (G3P)
enzymes by mutations that impair biosynthesis transforming competent
cells, in the presence of an auxotrophic
effect with these differences generated for an inability of the host
organism to synthesize an essential
compound during glycolysis in Tb-TIM. Trypanosomal-TIM is a
glycolytic enzyme essential for the parasite survival
that causes Chagas*
disease, in this study G.
bellum
from the genus related Geraniaceae and its phenolic compound are
leads which generates an unstable epimer
of an enzyme Geranin
A-containing changes resulting from ligand
adducts in the active site to capture in addition a source
of frustration
that becomes more favourable. Glycolaldehyde
(GA)
the simplest sugar-related molecules uptake of a proton by Glu167
preserves the small effect for inhibition by PGA (transition-state
analog) relative
to
substrate, G3P produces a triosephosphate isomerase with wild-type
activity, loop
6
adopts the "closed" desolvated
(+) conformation to facilitate
completion of catalysis by the formation of the › Michaelis-Menten
complex (on the ‹ micros-ms
› time scale) utilization yields further corrected calculations
with corresponding (slower Kcat) motional
rates*
Km. Increase's are discussed in the context of the significance
(Enzyme kinetics\Kcat) and may be estimated where the
'single-substrate' is locked in a protein cage probably because of an active
reaction site (loop
6)
movement to the transition state for deprotonation; which are the
on-average opened (substrate binding and release) and closed
(activated for catalysis) of both monomers optimal WT (wild type) TIM
conformations. Lys-12
‹ is expected to interact with both centers, where the enediol
intermediate along with the catalytic glutamate
base and histidine-95
the catalytic electrophile stabalizes the reversible
reaction intermediate that polarizes the substrate DHAP
in the Michaelis
complex. Interconversion spans the C-terminal
end of the eight
β-strands.
For catalysis to occur likley
a low pKa value transition from DHAP - for the enolase reaction
enzyme enhancement 'relative
to
the nonenzymatic reaction - (Bound PGH
- phosphoglycolohydroxamate mimics the (closed form) negative
polarization (•)
charge••,
while PGA
(2-phosphoglycolate) the positively charged residues in the two
active conformation sites.) is similar for the two conformers' in the
closed
conformation, on ligand
binding interacting with the reactive end's (β)
the deprotonated substrate-bound structures to be protonated by a
single-base (Glu-165)
proton transfer^ mechanism.

Structure of human triose phosphate isomerase at the positions of introns in homologous TPI genes from a number of phylogenetically diverse species. The introns motif are identified as calculated in phylogeny.
Phylogenetic trees constructed on the basis of sequence comparisons for triosephosphate isomerases analysis, TIM sequences were constructed based phylogeny with similarity, to those adopting the same structural fold of interest from different species for the taxonomic groups and the K13M mutations involvement in the human triosephosphate isomerase gene family...
Interactions
in the loop regions combine the effects of His95 and Lys13 for Glu165
(loop 4, 1,
and 6) the three crucial catalytic residues in triose phosphate
isomerase, all
form the enediol intermediate necessary for the interconversion reaction catalyzed by
TIM resulting in the natural substrates G3P formation. The introns
motif are identified as calculated in phylogenic
motifs.
Poorly conserved residues as targets for specific••
drug design are expected when compared to (TPI)
Triosephosphate isomerase (•). Catalytic residues of the
phylogenetic relationship pathways obtained by sequence based methods
of specific key amino acids can than be calculated to the incidence
residues and other TIMs which may influence the (human) HsTPI
equilibrium.