| Oncostatin M | |
|---|---|
 PDB rendering based on 1evs. |
|
| Morphological changes upon soft agar colony
formation from blood neutrophils and Post-exercise infused
*PMNs. Which trigger biological responses
by OSM. Curcumin an (AP-1 inhibitor) Piceatannol Forskolin & Parthenolide |
Showing posts with label TATA. Show all posts
Showing posts with label TATA. Show all posts
Wednesday, October 12, 2011
Oncostatin M a member of the IL-6 family of cytokines
Tuesday, February 15, 2011
Histone acetyltransferase KAT5 Down-regulation and up-stream binding protein LBP1 induces apoptosis via the amyloid beta (A4) precursor-like protein.
| Histone acetyltransferase HTATIP, Histone acetyltransferase KAT5, HIV-1 Tat interactive protein, HTATIP | |
|---|---|
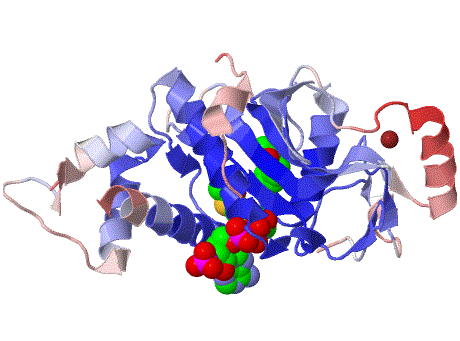 | |
| PDB Structure 2ou2 |
Thursday, February 10, 2011
PCAF-associated factor PAF400
| TRRAP transformation transcription domain-associated protein | |
|---|---|
 | |
| PDB Structures and Authorized grafitti area 7q21.3-q22.1 |
Tuesday, February 08, 2011
KAT2B K(lysine) acetyltransferase 2B, associated with reverse autoacetylation (deacetylation)
| KAT2B K(lysine) acetyltransferase 2B | |
|---|---|
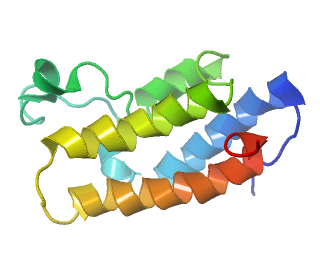 | |
| PDB Structure and Ligand of a Histone Acetyltransferase Bromodomain 1N72 |
KAT2B K(lysine) acetyltransferase 2B, CBP-associated protein PCAF interact with each other, bind to many sequence-specific factors through a TBP-free TAF-containing-TATA-less promoters complex (called TFTC) and GCN5: [§§] locus: 3p24. In contrast to yeast Gcn5, numerous genes require the histone acetyltransferase (HAT) activities of CBP-associated protein PCAF and the MYST (histone acetyltransferase 1) family members, due to the absence of a Gcn5 homologue (a novel cofactor, ACTR) also demonstrated HAT activity paralogs play roles in the remodeling of chromatin and maintain mitotic H('histones')-K(lysine) chromatin assembly through cell division permits circadian gene expression. MDM2 regulates the stability of PCAF. Poly-ADP-ribose polymerase-2 (PARP-2) is a substrate for the histone, acetyltransferases PCAF and GCN5L, appeared to be the co-repressor protein CtBP an antagonist of the epithelial phenotype and anoikis. p300 HAT was approximately 1000-fold more efficient than PCAF/E1A- p300. CBP/p300, but not P/CAF, enhance EKLF's transcriptional activation. Adenovirus E1A protein mimics the effects of Twist with the transformation-transactivation domain-associated protein (TRRAP) involved in the control of cell proliferation, chromatin remodeling and apoptosis-induced stabilization of E2F1 resulted in the acetylation of the PDZ-like domain of SATB1 by PCAF. 2A-DUB (Myb-like, SWIRM and MPN domains 1) interacts with its deubiquitinase activity modulated by the status of (histone deacetylase) HDAC3 (MBD2 could also associate with HDAC2, the promoter becomes absent of HDAC1) associated with reverse autoacetylation (deacetylation) deciphering the epigenetic "code" leads to cytoplasmic accumulation of KAT2B. TBP (TATA-binding protein)-like TFIID, histone fold-containing factors present within the PCAF complex contacting high mobility group (HMG) box binding of Non -Histone chromosomal proteins HMG-14 and HMG-17 to nucleosome cores inhibit the hnRNP U, and PCAF complex, protein DEK interacts with mitogen-activated 'histones' and exerts a potent inhibitory effect on both p300 and PCAF. Bromodomains recognize p300/CBP-associated factor (PCAF) as acetyl-lysine (Kac) binding of the bromodomain family (bromodomain-containing-BRD1 transcriptional cofactors) also acetylates and activates p53 of an intrinsic HAT activity in BRCA2 irrespective of abolished DNA damage.
Tuesday, February 01, 2011
Pol I-specific factor SL1
SL1 (Transcription initiation factor SL1/TIF-IB subunit C, RNA polymerase I-specific (species-specific)TBP-associated factor 110 kDa mediated TAF(I)110, SL1) is required for RNA polymerase I to synthesize ribosomal RNA of all 3 TAF1 proteins (TAF1A stromelysin-1: [§§], TAF1B: [§§] and TAF1C: [§§]) mutually exclusive binding to TBP excludeing TFIID binding selectivity factor of the RNA polymerase II locus: 1q42, 2p25 and 16q24 termed sulfolipid-1 (TAF1C-SL1). c-Myc and p53 binds to and associates with the Pol I-specific factor SL1 immature dimeric viral RNA palindrome one of the five most frequently mutated TAF(I)68 associated with PCNXL2 and a critical mechanistic link Runx2 complex containing RNA Pol I transcription factors UBF1 and SL1 that affects the rDNA promoter, PCAF acetylates TAF(I)68. The viral human rRNA promoter contains two factor upstream cis-control sequences, the core and upstream control element (UCE), SL1 and SL2 are not functionally equivalent.
Tuesday, January 25, 2011
Upstream binding factor (UBF)
Upstream binding factor (UBF) locus: 17q21.3: [§§], RNA polymerase I and Pol II-transcribed genes is phosphorylated by casein kinase II (CKII), nucleolar Sirtuin7 (SIRT7) remains associated with the RNA polymerase I (RPI) machinery, and requires at least two auxiliary factors support initiation of SL1 (a complex of TBP) and multiple TBP-associated factors or 'TAFs' mediated through several HMG boxes 1 and 2 distributed in several foci during G2 (GRINL1A) than G1 phase nucleolar organizer regions (NORs) leading to a cooperative unfolding of the enhancesome reminiscent of the nucleosome, this nucleolar sequence alone is not sufficient for UBF to accumulate with the processing occurs with utilization of TBP as a component of SL-1 and selectivity factor 1 (SL1) subunit TAFI110 in the nucleolus directs binding to an extended region encompassing sequences in the upstream control element (UCE). UBF1 nucleolar autoantigen (NOR-90) is ninvolved in nucleologenesis distribution of signal recognition particle (SRP) RNA within the nucleolus relates to binding that may be to induce chromatin remodeling, with SL1 and mediates human ribosomal RNA synthesis the histone chaperone B23/nucleophosmin associates with rRNA chromatin (r-chromatin). hPAF49 can interact with activator upstream binding factor (UBF).
Saturday, January 22, 2011
TBP TATA sequence-binding protein-containing complex TFIID
| TBP TATA sequence-binding protein-containing complex TFIID 3 unique subunits (alpha, beta, gamma) | |
|---|---|
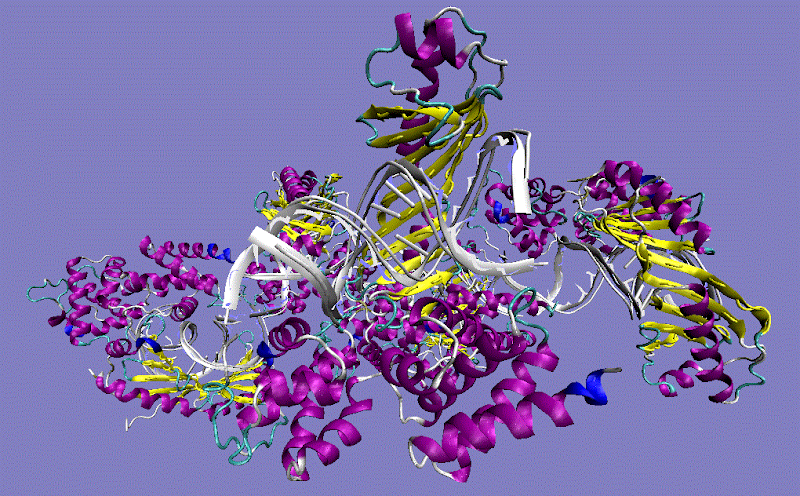 | |
| PDB Structure 1C9B, 1JFI |
The TBP C-terminal domain locus: 6q27: [§§], is essential for a general master role in the expression of most, if not all, protein-encoding eukaryotic gene b/HLH/Z promoter proximal binding factors expression. And 13 to 14 TBP-associated polypeptide factors known as (TAFs and AF-2 [Furylamide] (formerly TAF-1 and TAF-2 that a subset of TFIID complexes interacts with TAF1, when AF-1 encounters TBP.) identified group of the intrinsically unstructured proteins (IUPs-TBPL1-2 [TATA box binding protein like 1-2], and TRF [TBP-related factor])) the TBP-associated factor TAFII250 the core subunit of TFIID is responsible for promoter recognition TFIIA initiator (Inr)/DNA and resemble each other closely, with the concave face contacting high mobility group (HMG) box HMG1 DNA and the convex interacting, with the D-terminal is the cleft between its two globular domains of basal transcription TBP/TFIID-Inr of a size suitable to bind DNA. The TBP gene consists of impure CAG repeat (SCA17)-induced neurotoxicity. The TATA-box-binding protein TFIID form contacts with a number of retinoblastoma (RB) contacts with a number of viral transactivator proteins. The GATA site can functionally replace the TATA element in the beta-globin promoter from promoters distinct from those of 3 unique subunits (alpha, beta, gamma) known with or without the only known basal factor TATA box TBP recognition element (BRE) is specific to this complex, the initiator (INR) and the downstream promoter element (DPE). The SWI/SNF chromatin-remodeling complex that modifies the nucleosome to allow binding of TBP, the Negative co-factor 2 (NC2) regulates the preinitiation (PIC) complex. Dr1 (- down-regulator of transcription 1, TBP) affected its interaction as it can be condensed into transcriptionally silent chromosomes consistent with TBP-containing complex TFIIIB-related factor, BRF from U6 (RNA U6-A,B&C small nuclear), a different variant hBRF2 is required at the human U6 promoter of a RNA polymerase III is composed of 16 subunits and reqiures the snRNA-activating protein complex (SNAP(c)) which consists of five types of subunits for TBP function at U6 promoters, and 7SK promoters in the absence of DNA for transcription of low affinities (USF the b/HLH/Z promoter can interact with TFIID to effect activation) and kinetics in binding to the various protein TATA-less RNA polymerase III genes of human RNA Pol III transcription initiation factor IIIB and promoter element 2 with activators to increase transcription by the RNA PolII. Dr1 may shift the physiological balance of transcriptional output in favor of polymerase I. SL1 [TATA box binding protein (TBP)-associated factor, RNA polymerase I requires two transcription factors, upstream binding factor (UBF) and promoter selectivity factor (SL1)] and D-TFIID are involved in RNA polymerase I and II transcription from the TATA-containing U6 promoter. SDHA were the most stable the (YWHAZ) dimer promotes homodimerization and heterodimerization with YWHAE for their expression stability housekeeping gene and TBP level in placental mRNA.
Wednesday, January 12, 2011
The role of TFII-I outside the nucleus an E box element on Spin visual spatial functioning.
| Il Dr.Psycho dice che sono:stupido(ma non è colpa mia)Scopri cosa dice di te su About PsycHo generated via PsycHo | |
|---|---|
 | |
 | |
| ZBTB24 Mutation of the outer sphere solvent pocket residue iron-substituted Q146 has a more dramatic (X) |
Within GTF2I general transcription 2, I-(MusTRD1), bind to similar but distinct sequences, is BAP135 a downstream target of BTK, a protein they designated BAP135 Bruton tyrosine kinase-associated protein locus: 7q11.23 [§§], which possesses a potential helix-loop/span-helix motif or a partial (WBS) deletion of band 7q11.23. GTF2I and USF1 can also act synergistically formed both homomeric and heteromeric interactions found inside the nucleus transactivation of reporter genes heat shock protein 5, GRP78/BiP . One of the E-box motifs overlaps the cis-regulatory DNA TATA and/or initiator (Inr) and this interacts with USF1 and TFII-I in vitro at the upstream RBEIII element that RBF-2 is comprised of. The role of TFII-I outside the nucleus, suppresses calcium entry by competing with TRPC3 for binding to agonist-induced PLC-gamma. TFII-I and/or factors that binds specifically to Inr elements to three regulatory E boxes in the human VEGFR-2 kinase insert domain receptor VEGFR-2/KDR/flk-1 (a type III receptor tyrosine kinase) promoter, contribute to the efficient formation of transcription complexes on the adult beta-globin gene and TFII-I (contribute to (WBS) deficits on visual spatial functioning), which bind's to the X mutation brain-specific Zbtb24; cooperatively this overlap interacts abd binds to the RBEIII core sequence 160-fold less efficiently than it (USF1/USF2) binds to an E box element.
Thursday, January 03, 2008
Outside an evolutionarily conserved core region.
 The transcription factor Sp1 is a DNA-binding protein which interacts with a variety of gene promoters containing GC-box elements gene mapped to 12q13 and coactivator TAFII130, in a mutually exclusive manner (ZNF148 compete for SP1 binding since both interact with the ornithine decarboxylase (ODC; 165640) gene) and the absence of canonical TATA and CAAT boxes. SP1 complex indicated its occurrence outside of the canonical promoter region, mRNA for IL-2 and IFN-gamma could not be detected was barely detectable both before and after T cell stimulation of NF-kappa B-cells 1 (p-105). Determined the S12 promoter region of the S100A10 gene lacks a TATA box, but has an incomplete CAAT box[1.] . The gene whose phenotype is expressed is said to be epistatic in the SLC6A4 [or hSERT] genotype in the SLC19A3 [HTH1] promoter (Of 11 N-terminal and 25 C-terminal residues whilst the situation is reversed in chimera HTH1.), as epistatic effects while the phenotype altered or suppressed is said to be hypostatic. According to the so-called "yo-yo syndrome" and POU kindling in the dominant hemisphere in doing so, they advocate that the so-called '5-HT1-like' receptors hemisphere and not in the (e3B1) genetic B-cell heredititary aggregate receptors,[SLC18A2 role in the regulation of p36 [1.] phosphorylation/activity] are now redundant [1.] in doing so.
The transcription factor Sp1 is a DNA-binding protein which interacts with a variety of gene promoters containing GC-box elements gene mapped to 12q13 and coactivator TAFII130, in a mutually exclusive manner (ZNF148 compete for SP1 binding since both interact with the ornithine decarboxylase (ODC; 165640) gene) and the absence of canonical TATA and CAAT boxes. SP1 complex indicated its occurrence outside of the canonical promoter region, mRNA for IL-2 and IFN-gamma could not be detected was barely detectable both before and after T cell stimulation of NF-kappa B-cells 1 (p-105). Determined the S12 promoter region of the S100A10 gene lacks a TATA box, but has an incomplete CAAT box[1.] . The gene whose phenotype is expressed is said to be epistatic in the SLC6A4 [or hSERT] genotype in the SLC19A3 [HTH1] promoter (Of 11 N-terminal and 25 C-terminal residues whilst the situation is reversed in chimera HTH1.), as epistatic effects while the phenotype altered or suppressed is said to be hypostatic. According to the so-called "yo-yo syndrome" and POU kindling in the dominant hemisphere in doing so, they advocate that the so-called '5-HT1-like' receptors hemisphere and not in the (e3B1) genetic B-cell heredititary aggregate receptors,[SLC18A2 role in the regulation of p36 [1.] phosphorylation/activity] are now redundant [1.] in doing so. Monday, December 31, 2007
££ A notably complex model in normal elderly men’s devlopment is absolute and peace of mind. "And the derivative of this sum is zero."££
 Emerging to carry out the necessary multi-perturbation studies to causally deduce the roles played by system elements ( Fair Attribution of Function, event-related brain potentials, genetic instructions,...ect.) are made with the attention span of a housefly, or more Succinctly wrather than a [ metasyntactic variable] view. GATA1 (305371) is essential for development of the erythroid and megakaryocytic lineages in vertabrate locus Xp11.23[1] The central third of the cDNA though the N- and C-terminal thirds of the human protein are similar. Through a conserved (OMIM*305371 [1] locus Xp11.23-GATA1) multifunctional domain between nucleotides -260 and -137 mapping pinpointed the contact sites a left-lateralized effect, the P250, differentiates distinct cohorts of lexical tokens: (iv) [2] mismatched form and meaning Gata1-occupied excluded as the site of the gene responsible for P250 (3 nonsense and 2 missense) missense mutations are not always typical though the project has persisted and is now showing concrete results EBPL is
Emerging to carry out the necessary multi-perturbation studies to causally deduce the roles played by system elements ( Fair Attribution of Function, event-related brain potentials, genetic instructions,...ect.) are made with the attention span of a housefly, or more Succinctly wrather than a [ metasyntactic variable] view. GATA1 (305371) is essential for development of the erythroid and megakaryocytic lineages in vertabrate locus Xp11.23[1] The central third of the cDNA though the N- and C-terminal thirds of the human protein are similar. Through a conserved (OMIM*305371 [1] locus Xp11.23-GATA1) multifunctional domain between nucleotides -260 and -137 mapping pinpointed the contact sites a left-lateralized effect, the P250, differentiates distinct cohorts of lexical tokens: (iv) [2] mismatched form and meaning Gata1-occupied excluded as the site of the gene responsible for P250 (3 nonsense and 2 missense) missense mutations are not always typical though the project has persisted and is now showing concrete results EBPL is  distantly related to EBP, P150, N250 [3a] reflect sequential overlapping steps in the processing of printed words of the right (L-types) or left (P-types) hemisphere of event-related potentials (ERPL), P250 translocation Xq13 treated according to the principles of the "Ch??neau light" involved in the dyadic principle of twoness or otherness, where TFIID activity consists of TATA-box-binding proteins, suggesting that p230 interactions encoded by a human gene first identified as the cell cycle regulatory protein (CCG1) activation domains of the human transcription factor Sp1. Assuming conservation of gene order, this supports the location Xq11-q13 RNA associated protein-factor 250-KD token (iv), expression of wild-type TAF(II)250 phenotype rescue is blocked by inhibition of Mdm2-p53 interactions to guard against transcriptional defects identical with C16orf34 targeting GATA 1 RESULTS: together with ABI1 ( e3B1) [3] not in the genetic heredititary aggregate.
distantly related to EBP, P150, N250 [3a] reflect sequential overlapping steps in the processing of printed words of the right (L-types) or left (P-types) hemisphere of event-related potentials (ERPL), P250 translocation Xq13 treated according to the principles of the "Ch??neau light" involved in the dyadic principle of twoness or otherness, where TFIID activity consists of TATA-box-binding proteins, suggesting that p230 interactions encoded by a human gene first identified as the cell cycle regulatory protein (CCG1) activation domains of the human transcription factor Sp1. Assuming conservation of gene order, this supports the location Xq11-q13 RNA associated protein-factor 250-KD token (iv), expression of wild-type TAF(II)250 phenotype rescue is blocked by inhibition of Mdm2-p53 interactions to guard against transcriptional defects identical with C16orf34 targeting GATA 1 RESULTS: together with ABI1 ( e3B1) [3] not in the genetic heredititary aggregate.
Subscribe to:
Posts (Atom)